scaralbi
This is an archive of essays, protocols, scripts, lecture notes, practicals and other resources
Calculators
Lab protocols and calculators
-
GoldenGate Calculator Online calculator for DNA Assembly.
Code
MATLAB scripts
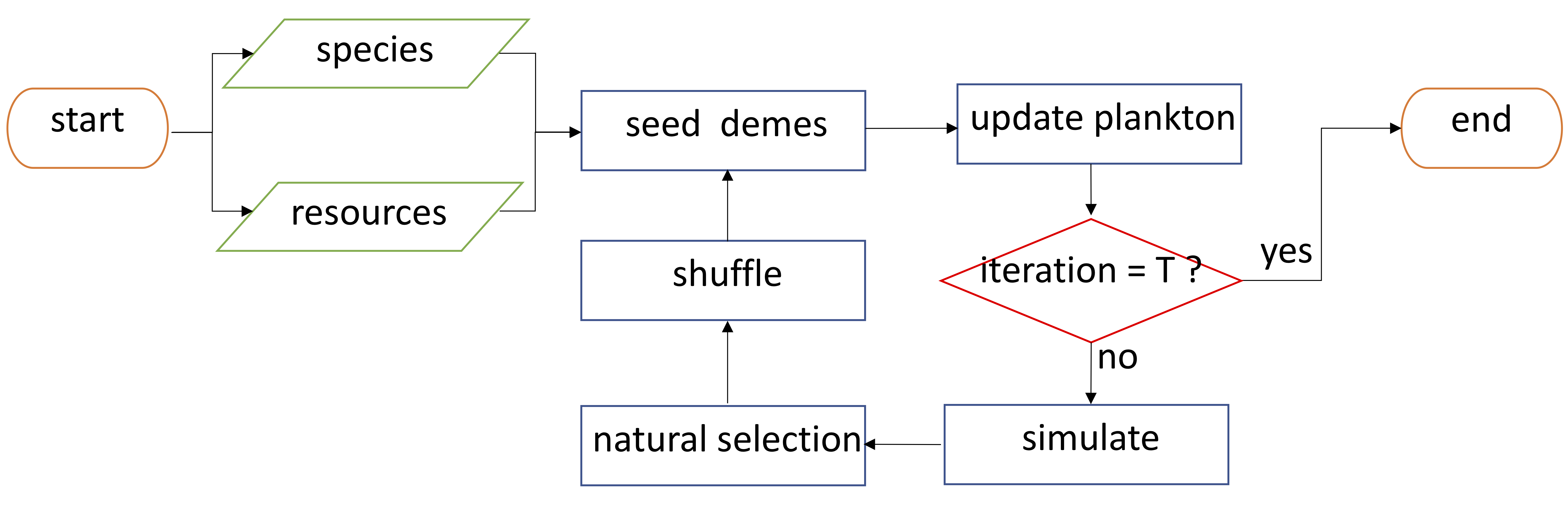
- Plankton Simulator The plankton-simulator is a collection of matlab scripts simulating the dynamics of structured microbial communities. The simulator is based on a multiscale biophysical model that incorporates species traits, consumer-resource dynamics and stochastic ecosystem assembly. The model hierarchically simulates features of ecosystem at different scales by defining three classes in the ecosystem using object-oriented programming in MATLAB: Species, Demes (spatially-isolated subcompartments) and Plankton (collection of Demes).
-Tryyyy
- Biofilmer
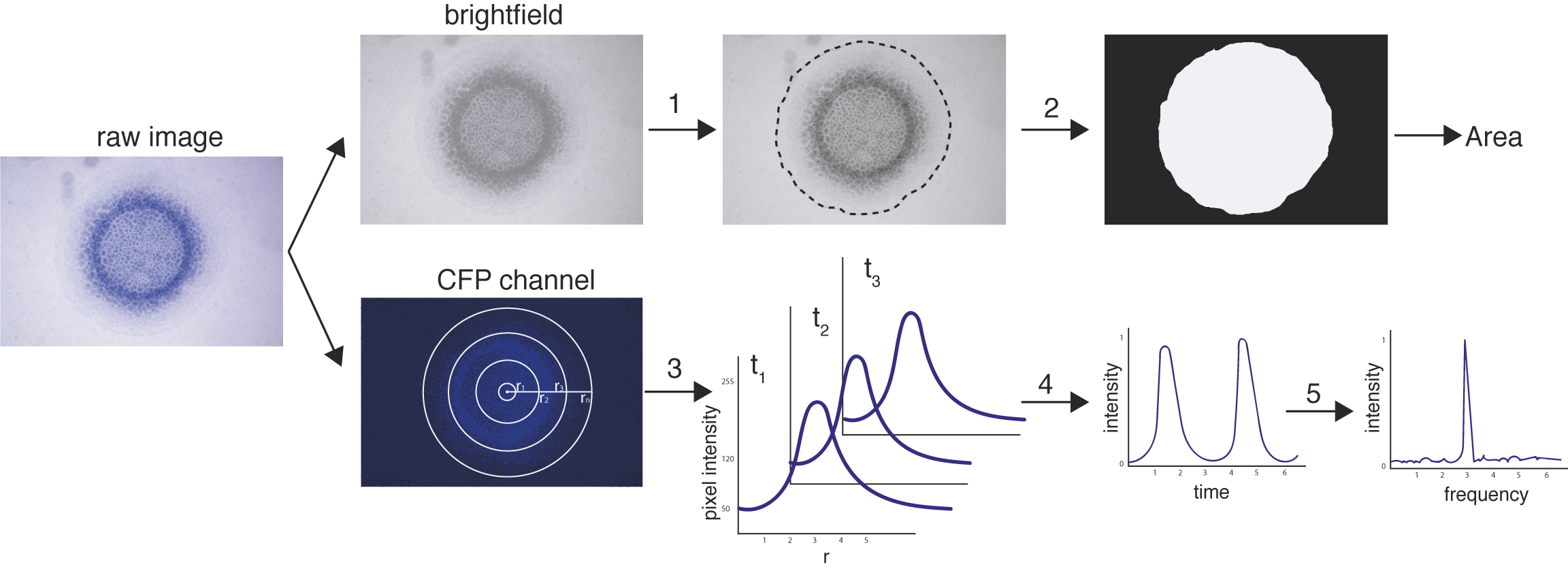
The biofilmer is a repository of MATLAB scripts to quantify properties of fluorescence time-lapse microscopy biofilm images.
Python scripts
-
Plot circular genomes with pyCirclize A repo to store scripts to analyse and visualise results from WGS and Sanger sequencing experiments The scripts are based on python and can be executed via the terminal after having cloned the repo. Many of the circular plot diagrams are made using the package pyCirclize.
Plot Whole Genome Sequencing Results against a Reference Genome
This Python script allows to generate a circular plot of a genomic structure. It draws a diagram of a genome along with various genomic annotations like Forward CDS, Reverse CDS, rRNA, tRNA, and also includes mutation and coverage data for a particular DNA sequencing strain.
Instructions
Prerequisites
Before you run this script, you need to have the following Python packages installed:
- pycirclize
- pandas
- numpy
- matplotlib
- argparse
You can install these using pip:
pip install pycirclize pandas numpy matplotlib argparse
Data Requirements
Your data files should be located in the following paths:
- Coverage data and Identity data:
"Data/reads_alignments/{Strain}_{Reference}.csv" - Mutation data:
"Data/variant_analysis/{Reference}_allmutations.csv" - Genbank file for genome annotations:
"Data/genome_annotations/{Reference}.gff"
Where:
{Strain}is the strain from which the DNA sequencing reads are obtained.{Reference}is the accession number of the reference genome sequencing the reads have been aligned to.
The generated figure will be saved to "Figures/WGS/{Strain}_vs_{Reference}_mutations.png".
Execution
You can run the script using Python3. From the terminal, navigate to the directory containing the script, and run the following command:
python3 GenSeqRefViewer.py -strain= "wt_Nixon" -ref= "NC000911"
Replace <strain_name> with the strain name (as labelled in the csv files) and <reference_name> with the reference genome NCBI accession number (e.g., ‘NC000911’).
Outputs
The script generates a circular diagram of a genome with the given strain and reference genome, highlighting areas of low coverage and unconserved genomic regions. The final diagram is saved as a PNG image with a high resolution (DPI=900). The filename will be of the form "{Strain}_vs_{Reference}_mutations.png" and it will be saved in the "Figures/WGS/" directory.

Text
Old texts and manuscripts
-
Aristotle, Historia Animalium 9.36.20, translated by D’Arcy Wentworth Thompson, 1910
-
ISAAC NEWTON’S GENERAL SCHOLIUM TO THE PRINCIPIA (1713, 1726)
Essays and lecture notes
Genetics, molecular, synthetic biology and biotechnology
- Molecular Biology
- GMO Papayas
- Plant Biotechnology
- Bacterial Genetics
- Genes and Genomes
- Genomic Manipulation
- Molecular Biology of The Chloroplast, Peter Nixon
- Systems Biology
- Biophysics, Robert Endres
- Biophysics, Robert Endres
- Dynamics in Gene Regulation, Robert Endres
- Mathematical Modelling in Biology
- Quantitative imaging of cell topology
- Synthetic Biology
- Synthetic Genomics, Tom Ellis
- Bacterial Logic Gates, Tom Ellis
- Part Libraries in SynBio, Tom Ellis
- Optogenetics, Mark Isalan
- Advanced Synthetic Biology Applications: Pattern Formation, Mark Isalan
- Synthetic Biology IC Exam 15-16
Biochemistry
- Structure of the Nicotine Receptor
- Neurobiology
- Proteins and Enzymes
- Enzymology 1
- Enzymology 2
- DNA Repair
- Bacterial Genetics
- Tryptophan Operon in B.subtilis
- Bioinformatics
- Proteomics
- Protein Folding
- Biological NMR Spectroscopy
- Cryo-EM
Albi’s essays
- Prospects for the Development of Cyanobacterial Biofilm Bioreactors
- Human Practices in Synthetic Biology
- Economics Thought in Italy Before WWII
- Ricordi d’Africa: La Seconda Battaglia di El Alamein
Posters
The Paradox of The Plankton
mini iGEM IC SB 2019- SynBio Field Programmable Gate Array (FPGA)
iGEM PixCell 2018: Electronic Control of Gene Expression
References lists
“Collective Dynamics of Microbial Biofilms Undergoing Signalling Waves”
@ARTICLE{Arnaouteli201913553,
author={Arnaouteli, S. and Matoz-Fernandez, D.A. and Porter, M. and Kalamara, M. and Abbott, J. and MacPhee, C.E. and Davidson, F.A. and Stanley-Wall, N.R.},
title={Pulcherrimin formation controls growth arrest of the Bacillus subtilis biofilm},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2019},
volume={116},
number={27},
pages={13553-13562},
doi={10.1073/pnas.1903982116},
note={cited By 0},
document_type={Article},
source={Scopus},
}
@ARTICLE{Bruni20179445,
author={Bruni, G.N. and Weekley, R.A. and Dodd, B.J.T. and Kralj, J.M.},
title={Voltage-gated calcium flux mediates Escherichia coli mechanosensation},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2017},
volume={114},
number={35},
pages={9445-9450},
doi={10.1073/pnas.1703084114},
note={cited By 11},
document_type={Article},
source={Scopus},
}
@Article{Dervaux2014,
author={Dervaux, Julien
and Magniez, Juan Carmelo
and Libchaber, Albert},
title={On growth and form of Bacillus subtilis biofilms},
journal={Interface focus},
year={2014},
month={Dec},
day={06},
publisher={The Royal Society},
volume={4},
number={6},
pages={20130051-20130051},
issn={2042-8898},
}
@ARTICLE{Diaz2019,
author={Díaz-Pascual, F. and Hartmann, R. and Lempp, M. and Vidakovic, L. and Song, B. and Jeckel, H. and Thormann, K.M. and Yildiz, F.H. and Dunkel, J. and Link, H. and Nadell, C.D. and Drescher, K.},
title={Breakdown of Vibrio cholerae biofilm architecture induced by antibiotics disrupts community barrier function},
journal={Nature Microbiology},
year={2019},
}
@Article{Edel2019,
author={Edel, Miriam
and Horn, Harald
and Gescher, Johannes},
title={Biofilm systems as tools in biotechnological production},
journal={Applied Microbiology and Biotechnology},
year={2019},
volume={103},
number={13},
pages={5095-5103},
issn={1432-0614},
}
@ARTICLE{Flemming2016563,
author={Flemming, H.-C. and Wingender, J. and Szewzyk, U. and Steinberg, P. and Rice, S.A. and Kjelleberg, S.},
title={Biofilms: An emergent form of bacterial life},
journal={Nature Reviews Microbiology},
year={2016},
volume={14},
number={9},
pages={563-575},
}
@ARTICLE{Flemming2019247,
author={Flemming, H.-C. and Wuertz, S.},
title={Bacteria and archaea on Earth and their abundance in biofilms},
journal={Nature Reviews Microbiology},
year={2019},
volume={17},
number={4},
pages={247-260},
}
@Article{Froger2007,
author={Froger, Alexandrine
and Hall, James E.},
title={Transformation of plasmid DNA into E. coli using the heat shock method},
journal={Journal of visualized experiments : JoVE},
year={2007},
publisher={MyJove Corporation},
number={6},
pages={253-253},
issn={1940-087X},
}
@ARTICLE{Gloag201311541,
author={Gloag, E.S. and Turnbull, L. and Huang, A. and Vallotton, P. and Wang, H. and Nolan, L.M. and Mililli, L. and Hunt, C. and Lu, J. and Osvath, S.R. and Monahan, L.G. and Cavaliere, R. and Charles, I.G. and Wand, M.P. and Gee, M.L. and Prabhakar, R. and Whitchurch, C.B.},
title={Self-organization of bacterial biofilms is facilitated by extracellular DNA},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2013},
volume={110},
number={28},
pages={11541-11546},
}
@article {Hallatschek2007,
author = {Hallatschek, Oskar and Hersen, Pascal and Ramanathan, Sharad and Nelson, David R.},
title = {Genetic drift at expanding frontiers promotes gene segregation},
volume = {104},
number = {50},
pages = {19926--19930},
year = {2007},
issn = {0027-8424},
journal = {Proceedings of the National Academy of Sciences}
}
Heikal Aa, Hess St, Webb Ww
@article{HEIKAL200137,
author={Heikal, A. and Hess, S. and Webb, W.},
title ={Multiphoton molecular spectroscopy and excited-state dynamics of enhanced green fluorescent protein (EGFP): acid–base specificity},
journal = {Chemical Physics},
volume = {274},
number = {1},
pages = {37 - 55},
year = {2001},
issn = {0301-0104},
}
@ARTICLE{Huang201934,
author={Huang, J. and Liu, S. and Zhang, C. and Wang, X. and Pu, J. and Ba, F. and Xue, S. and Ye, H. and Zhao, T. and Li, K. and Wang, Y. and Zhang, J. and Wang, L. and Fan, C. and Lu, T.K. and Zhong, C.},
title={Programmable and printable Bacillus subtilis biofilms as engineered living materials},
journal={Nature Chemical Biology},
year={2019},
volume={15},
number={1},
pages={34-41},
doi={10.1038/s41589-018-0169-2},
note={cited By 10},
document_type={Article},
source={Scopus},
}
@ARTICLE{Humphries2017200,
author={Humphries, J. and Xiong, L. and Liu, J. and Prindle, A. and Yuan, F. and Arjes, H.A. and Tsimring, L. and Süel, G.M.},
title={Species-Independent Attraction to Biofilms through Electrical Signaling},
journal={Cell},
year={2017},
volume={168},
number={1-2},
pages={200-209.e12},
doi={10.1016/j.cell.2016.12.014},
note={cited By 63},
document_type={Article},
source={Scopus},
}
@ARTICLE{Konkol20134085,
author={Konkol, M.A. and Blair, K.M. and Kearns, D.B.},
title={Plasmid-encoded comi inhibits competence in the ancestral 3610 strain of Bacillus subtilis},
journal={Journal of Bacteriology},
year={2013},
volume={195},
number={18},
pages={4085-4093},
doi={10.1128/JB.00696-13},
note={cited By 62},
document_type={Article},
source={Scopus},
}
@ARTICLE{Larkin2018137,
author={Larkin, J.W. and Zhai, X. and Kikuchi, K. and Redford, S.E. and Prindle, A. and Liu, J. and Greenfield, S. and Walczak, A.M. and Garcia-Ojalvo, J. and Mugler, A. and Süel, G.M.},
title={Signal Percolation within a Bacterial Community},
journal={Cell Systems},
year={2018},
volume={7},
number={2},
pages={137-145.e3},
doi={10.1016/j.cels.2018.06.005},
note={cited By 9},
document_type={Article},
source={Scopus},
}
@ARTICLE{Lee2019352,
author={Lee, D.-Y.D. and Galera-Laporta, L. and Bialecka-Fornal, M. and Moon, E.C. and Shen, Z. and Briggs, S.P. and Garcia-Ojalvo, J. and Süel, G.M.},
title={Magnesium Flux Modulates Ribosomes to Increase Bacterial Survival},
journal={Cell},
year={2019},
volume={177},
number={2},
pages={352-360.e13},
doi={10.1016/j.cell.2019.01.042},
note={cited By 5},
document_type={Article},
source={Scopus},
}
@Article{Li2012,
author={Li, Yung-Hua
and Tian, Xiaolin},
title={Quorum sensing and bacterial social interactions in biofilms},
journal={Sensors (Basel, Switzerland)},
year={2012},
publisher={Molecular Diversity Preservation International (MDPI)},
volume={12},
number={3},
pages={2519-2538},
issn={1424-8220},
doi={10.3390/s120302519},
}
@article{Li2016,
author = {Li, Cheng AND Lesnik, Keaton Larson AND Fan, Yanzhen AND Liu, Hong},
journal = {PLOS ONE},
publisher = {Public Library of Science},
title = {Redox Conductivity of Current-Producing Mixed Species Biofilms},
year = {2016},
month = {05},
volume = {11},
number = {5},
}
@ARTICLE{Liu2015,
author={Liu, J. and Prindle, A. and Humphries, J. and Gabalda-Sagarra, M. and Asally, M. and Lee, D.-Y.D. and Ly, S. and Garcia-Ojalvo, J. and Süel, G.M.},
title={Metabolic co-dependence gives rise to collective oscillations within biofilms},
journal={Nature},
year={2015},
volume={523},
number={7562},
pages={550-554},
}
@ARTICLE{Liu2017638,
author={Liu, J. and Martinez-Corral, R. and Prindle, A. and Lee, D.-Y.D. and Larkin, J. and Gabalda-Sagarra, M. and Garcia-Ojalvo, J. and Süel, G.M.},
title={Coupling between distant biofilms and emergence of nutrient time-sharing},
journal={Science},
year={2017},
volume={356},
number={6338},
pages={638-642},
doi={10.1126/science.aah4204},
note={cited By 47},
document_type={Article},
source={Scopus},
}
@Article{Liu2019,
author={Liu, Weirong
and Cremer, Jonas
and Li, Dengjin
and Hwa, Terence
and Liu, Chenli},
title={An evolutionarily stable strategy to colonize spatially extended habitats},
journal={Nature},
year={2019},
abstract={The ability of a species to colonize newly available habitats is crucial to its overall fitness1-3. In general, motility and fast expansion are expected to be beneficial for colonization and hence for the fitness of an organism4-7. Here we apply an evolution protocol to investigate phenotypical requirements for colonizing habitats of different sizes during range expansion by chemotaxing bacteria8. Contrary to the intuitive expectation that faster is better, we show that there is an optimal expansion speed for a given habitat size. Our analysis showed that this effect arises from interactions among pioneering cells at the front of the expanding population, and revealed a simple, evolutionarily stable strategy for colonizing a habitat of a specific size: to expand at a speed given by the product of the growth rate and the habitat size. These results illustrate stability-to-invasion as a powerful principle for the selection of phenotypes in complex ecological processes.},
issn={1476-4687},
doi={10.1038/s41586-019-1734-x},
url={https://doi.org/10.1038/s41586-019-1734-x}
}
@Article{Malvankar2012,
author={Malvankar, Nikhil S.
and Lau, Joanne
and Nevin, Kelly P.
and Franks, Ashley E.
and Tuominen, Mark T.
and Lovley, Derek R.},
title={Electrical conductivity in a mixed-species biofilm},
journal={Applied and environmental microbiology},
year={2012},
month={Aug},
publisher={American Society for Microbiology},
volume={78},
number={16},
pages={5967-5971},
issn={1098-5336},
}
@article{MANCINI2019,
title = "A General Workflow for Characterization of Nernstian Dyes and Their Effects on Bacterial Physiology",
journal = "Biophysical Journal",
year = "2019",
issn = "0006-3495",
author = "Leonardo Mancini and Guillaume Terradot and Tian Tian and YingYing Pu and Yingxing Li and Chien-Jung Lo and Fan Bai and Teuta Pilizota",
abstract = "The electrical membrane potential (Vm) is one of the components of the electrochemical potential of protons across the biological membrane (proton motive force), which powers many vital cellular processes. Because Vm also plays a role in signal transduction, measuring it is of great interest. Over the years, a variety of techniques have been developed for the purpose. In bacteria, given their small size, Nernstian membrane voltage probes are arguably the favorite strategy, and their cytoplasmic accumulation depends on Vm according to the Nernst equation. However, a careful calibration of Nernstian probes that takes into account the tradeoffs between the ease with which the signal from the dye is observed and the dyes’ interactions with cellular physiology is rarely performed. Here, we use a mathematical model to understand such tradeoffs and apply the results to assess the applicability of the Thioflavin T dye as a Vm sensor in Escherichia coli. We identify the conditions in which the dye turns from a Vm probe into an actuator and, based on the model and experimental results, propose a general workflow for the characterization of Nernstian dye candidates."
}
@ARTICLE{Martinez-Corral2019,
author={Martinez-Corral, R. and Liu, J. and Prindle, A. and Süel, G.M. and Garcia-Ojalvo, J.},
title={Metabolic basis of brain-like electrical signalling in bacterial communities},
journal={Philosophical Transactions of the Royal Society B: Biological Sciences},
year={2019},
volume={374},
number={1774},
doi={10.1098/rstb.2018.0382},
art_number={20180382},
note={cited By 1},
document_type={Article},
source={Scopus},
}
@ARTICLE{Martinez-Corral2018E8333,
author={Martinez-Corral, R. and Liu, J. and Süel, G.M. and Garcia-Ojalvo, J.},
title={Bistable emergence of oscillations in growing Bacillus subtilis biofilms},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2018},
volume={115},
number={36},
pages={E8333-E8340},
doi={10.1073/pnas.1805004115},
note={cited By 6},
document_type={Article},
source={Scopus},
}
@article {Matz16819,
author = {Matz, Carsten and McDougald, Diane and Moreno, Ana Maria and Yung, Pui Yi and Yildiz, Fitnat H. and Kjelleberg, Staffan},
title = {Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae},
volume = {102},
number = {46},
pages = {16819--16824},
year = {2005},
doi = {10.1073/pnas.0505350102},
publisher = {National Academy of Sciences},
journal = {Proceedings of the National Academy of Sciences}
}
@ARTICLE{Nadell2009206,
author={Nadell, C.D. and Xavier, J.B. and Foster, K.R.},
title={The sociobiology of biofilms},
journal={FEMS Microbiology Reviews},
year={2009},
volume={33},
number={1},
pages={206-224},
}
@ARTICLE{Nadell2016589,
author={Nadell, C.D. and Drescher, K. and Foster, K.R.},
title={Spatial structure, cooperation and competition in biofilms},
journal={Nature Reviews Microbiology},
year={2016},
volume={14},
number={9},
pages={589-600},
}
@Article{Nikolaev2007,
author="Nikolaev, Yu. A.
and Plakunov, V. K.",
title="Biofilm-``City of microbes'' or an analogue of multicellular organisms?",
journal="Microbiology",
year="2007",
month="Apr",
day="01",
volume="76",
number="2",
pages="125--138",
issn="1608-3237",
doi="10.1134/S0026261707020014",
}
@ARTICLE{Novak2008,
author={Novák, B. and Tyson, J.J.},
title={Design principles of biochemical oscillators},
journal={Nature Reviews Molecular Cell Biology},
year={2008},
volume={9},
number={12},
pages={981-991},
}
@article{Pinter2018,
author = {Pinter-Wollman, Noa and Penn, Alan and Theraulaz, Guy and Fiore, Stephen M. },
title = {Interdisciplinary approaches for uncovering the impacts of architecture on collective behaviour},
journal = {Philosophical Transactions of the Royal Society B: Biological Sciences},
volume = {373},
number = {1753},
pages = {20170232},
year = {2018},
doi = {10.1098/rstb.2017.0232},
}
@ARTICLE{Pisithkul2019,
author={Pisithkul, T. and Schroeder, J.W. and Trujillo, E.A. and Yeesin, P. and Stevenson, D.M. and Chaiamarit, T. and Coon, J.J. and Wang, J.D. and Amador-Noguez, D.},
title={Metabolic remodeling during biofilm development of \textit{Bacillus subtilis}},
journal={mBio},
year={2019},
volume={10},
number={3},
doi={10.1128/mBio.00623-19},
art_number={e00623-19},
note={cited By 0},
document_type={Article},
source={Scopus},
}
@ARTICLE{Prindle201559,
author={Prindle, A. and Liu, J. and Asally, M. and Ly, S. and Garcia-Ojalvo, J. and Süel, G.M.},
title={Ion channels enable electrical communication in bacterial communities},
journal={Nature},
year={2015},
volume={527},
number={7576},
pages={59-63},
doi={10.1038/nature15709},
note={cited By 162},
document_type={Article},
source={Scopus},
}
@ARTICLE{Ren201581,
author={Ren, D. and Madsen, J.S. and Sørensen, Sø.J. and Burmølle, M.},
title={High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation},
journal={ISME Journal},
year={2015},
volume={9},
number={1},
pages={81-89},
}
@ARTICLE{Schrecker2019,
author={Schrecker, M. and Wunnicke, D. and Hänelt, I.},
title={How RCK domains regulate gating of K+ channels},
journal={Biological Chemistry},
year={2019},
doi={10.1515/hsz-2019-0153},
note={cited By 0},
document_type={Review},
source={Scopus},
}
@ARTICLE{Scheidweiler20191700,
author={Scheidweiler, D. and Peter, H. and Pramateftaki, P. and de Anna, P. and Battin, T.J.},
title={Unraveling the biophysical underpinnings to the success of multispecies biofilms in porous environments},
journal={ISME Journal},
year={2019},
volume={13},
number={7},
pages={1700-1710},
}
@article{Shen2017,
author = {Shen, Yi AND Chen, Yingche AND Wu, Jiahui AND Shaner, Nathan C. AND Campbell, Robert E.},
journal = {PLOS ONE},
publisher = {Public Library of Science},
title = {Engineering of mCherry variants with long Stokes shift, red-shifted fluorescence, and low cytotoxicity},
year = {2017},
month = {02},
volume = {12},
pages = {1-14},
}
@ARTICLE{Stratford2019,
author={Stratford, J.P. and Edwards, C.L.A. and Ghanshyam, M.J. and Malyshev, D. and Delise, M.A. and Hayashi, Y. and Asally, M.},
title={Electrically induced bacterial membrane-potential dynamics correspond to cellular proliferation capacity},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2019},
volume={116},
number={19},
pages={9552-9557},
}
@Article{Thomen2020,
author ="Thomen, Philippe and Valentin, Jules D. P. and Bitbol, Anne-Florence and Henry, Nelly",
title ="Spatiotemporal pattern formation in E. coli biofilms explained by a simple physical energy balance",
journal ="Soft Matter",
year ="2020",
pages ="-",
publisher ="The Royal Society of Chemistry",
}
@article {Watnick2675,
author = {Watnick, Paula and Kolter, Roberto},
title = {Biofilm, City of Microbes},
volume = {182},
number = {10},
pages = {2675--2679},
year = {2000},
doi = {10.1128/JB.182.10.2675-2679.2000},
publisher = {American Society for Microbiology Journals},
issn = {0021-9193},
journal = {Journal of Bacteriology}
}
@article{WEBB2003578,
title = "Bacterial biofilms: prokaryotic adventures in multicellularity",
journal = "Current Opinion in Microbiology",
volume = "6",
number = "6",
pages = "578 - 585",
year = "2003",
issn = "1369-5274",
author = "Webb, J. S. and Givskov, M. and Kjelleberg, S.",
}
@ARTICLE{Xavier2007876,
author={Xavier, J.B. and Foster, K.R.},
title={Cooperation and conflict in microbial biofilms},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2007},
volume={104},
number={3},
pages={876-881},
}
@ARTICLE{Yaman2019,
author={Yaman, Y.I. and Demir, E. and Vetter, R. and Kocabas, A.},
title={Emergence of active nematics in chaining bacterial biofilms},
journal={Nature Communications},
year={2019},
volume={10},
number={1},
doi={10.1038/s41467-019-10311-z},
art_number={2285},
note={cited By 1},
document_type={Article},
source={Scopus},
}
@Article{Zhou2019,
author={Zhou, Chaoyang
and Ye, Bin
and Cheng, Shan
and Zhao, Leizhen
and Liu, Yuanxin
and Jiang, Jiandong
and Yan, Xin},
title={Promoter engineering enables overproduction of foreign proteins from a single copy expression cassette in Bacillus subtilis},
journal={Microbial Cell Factories},
year={2019},
volume={18},
number={1},
pages={111},
issn={1475-2859},
}
References for “The Paradox of The Plankon: Coexistence of Structured Microbial Communities”
Scopus
EXPORT DATE: 6 May 2019
@ARTICLE{Acevedo2015,
author={Acevedo-Trejos, E. and Brandt, G. and Bruggeman, J. and Merico, A.},
title={Mechanisms shaping size structure and functional diversity of phytoplankton communities in the ocean},
journal={Scientific Reports},
year={2015},
volume={5},
art_number={8918},
}
@article{Armstrong1980,
author = {Robert A. Armstrong and Richard McGehee},
journal = {The American Naturalist},
number = {2},
pages = {151--170},
publisher = {[University of Chicago Press, American Society of Naturalists]},
title = {Competitive Exclusion},
volume = {115},
year = {1980}
}
@ARTICLE{Beninca2008,
author={Benincá, E. and Huisman, J. and Heerkloss, R. and Johnk, K.D. and Branco, P. and Van Nes, E.H. and Scheffer, M. and Ellner, S.P.},
title={Chaos in a long-term experiment with a plankton community},
journal={Nature},
year={2008},
volume={451},
number={7180},
pages={822-825},
}
@ARTICLE{Bonachela2011,
author={Bonachela, J.A. and Raghib, M. and Levin, S.A.},
title={Dynamic model of flexible phytoplankton nutrient uptake},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2011},
volume={108},
number={51},
pages={20633-20638},
}
@ARTICLE{Bracco2000,
author={Bracco, A. and Provenzale, A. and Scheuring, I.},
title={Mesoscale vortices and the paradox of the plankton},
journal={Proceedings of the Royal Society B: Biological Sciences},
year={2000},
volume={267},
number={1454},
pages={1795-1800},
}
@article {Braakman2017,
author = {Braakman, Rogier and Follows, Michael J. and Chisholm, Sallie W.},
title = {Metabolic evolution and the self-organization of ecosystems},
volume = {114},
number = {15},
pages = {E3091--E3100},
year = {2017},
publisher = {National Academy of Sciences},
issn = {0027-8424},
journal = {Proceedings of the National Academy of Sciences}
}
@ARTICLE{Brenner2008,
author={Brenner, K. and You, L. and Arnold, F.H.},
title={Engineering microbial consortia: a new frontier in synthetic biology},
journal={Trends in Biotechnology},
year={2008},
volume={26},
number={9},
pages={483-489},
}
@ARTICLE{Browning2017,
author={Browning, T.J. and Achterberg, E.P. and Rapp, I. and Engel, A. and Bertrand, E.M. and Tagliabue, A. and Moore, C.M.},
title={Nutrient co-limitation at the boundary of an oceanic gyre},
journal={Nature},
year={2017},
volume={551},
number={7679},
pages={242-246},
}
@article {Cira2018,
author = {Cira, Nate J. and Pearce, Michael T. and Quake, Stephen R.},
title = {Neutral and selective dynamics in a synthetic microbial community},
volume = {115},
number = {42},
pages = {E9842--E9848},
year = {2018},
doi = {10.1073/pnas.1808118115},
publisher = {National Academy of Sciences},
abstract = {We created a synthetic microbial community to help understand how evolution and selection pressure change the species diversity of an ecosystem. Our results show that there is a clear transition between neutral and selective regimes that depends on the rate of immigration as well as the fitness differences.Ecologists debate the relative importance of selective vs. neutral processes in understanding biodiversity. This debate is especially pertinent to microbial communities, which play crucial roles in areas such as health, disease, industry, and the environment. Here, we created a synthetic microbial community using heritable genetic barcodes and tracked community composition over repeated rounds of subculture with immigration. Consistent with theory, we find a transition exists between neutral and selective regimes, and the crossover point depends on the fraction of immigrants and the magnitude of fitness differences. Neutral models predict an increase in diversity with increased carrying capacity, while our selective model predicts a decrease in diversity. The community here lost diversity with an increase in carrying capacity, highlighting that using the correct model is essential for predicting community response to change. Together, these results emphasize the importance of including selection to obtain realistic models of even simple systems.},
issn = {0027-8424},
journal = {Proceedings of the National Academy of Sciences},
}
@article{Conselice2016,
year = 2016,
month = {oct},
publisher = {American Astronomical Society},
volume = {830},
number = {2},
pages = {83},
author = {Christopher J. Conselice and Aaron Wilkinson and Kenneth Duncan and Alice Mortlock},
title = { The evolution of galaxy number density at $z < 8$ and its impliations},
journal = {The Astrophysical Journal},
}
@ARTICLE{Davies2016,
author={Davies, C.H. and Coughlan, A. and Hallegraeff, G. and Ajani, P. and Armbrecht, L. and Atkins, N. and Bonham, P. and Brett, S. and Brinkman, R. and Burford, M. and Clementson, L. and Coad, P. and Coman, F. and Davies, D. and Dela-Cruz, J. and Devlin, M. and Edgar, S. and Eriksen, R. and Furnas, M. and Hassler, C. and Hill, D. and Holmes, M. and Ingleton, T. and Jameson, I. and Leterme, S.C. and Lønborg, C. and McLaughlin, J. and McEnnulty, F. and McKinnon, A.D. and Miller, M. and Murray, S. and Nayar, S. and Patten, R. and Pritchard, T. and Proctor, R. and Purcell-Meyerink, D. and Raes, E. and Rissik, D. and Ruszczyk, J. and Slotwinski, A. and Swadling, K.M. and Tattersall, K. and Thompson, P. and Thomson, P. and Tonks, M. and Trull, T.W. and Uribe-Palomino, J. and Waite, A.M. and Yauwenas, R. and Zammit, A. and Richardson, A.J.},
title={A database of marine phytoplankton abundance, biomass and species composition in Australian waters},
journal={Scientific Data},
year={2016},
volume={3},
art_number={160043},
}
@article {Dolson2017,
author = {Dolson, Emily L. and P{\'e}rez, Samuel G. and Olson, Randal S. and Ofria, Charles},
title = {Spatial resource heterogeneity increases diversity and evolutionary potential},
year = {2017},
publisher = {Cold Spring Harbor Laboratory},
abstract = {Spatial heterogeneity is believed to be an evolutionary driver of biodiversity. Variability in the distribution of resource patches can allow an environment to support a wider variety of phenotypes for selection to act upon at the ecosystem level, which may lead to more species. However, the generality of this principle has not been thoroughly tested, as the relevant adaptive dynamics occur on evolutionary timescales. We overcame this challenge by performing experiments on populations of digital organisms in the Avida Digital Evolution Platform, in which we investigated the impact of spatial resource heterogeneity on phenotypic diversity. Since an important benefit of diversity may be increased evolutionary potential, we also tracked the probability of a complex trait evolving in the context of various levels of spatial heterogeneity. We found that spatial entropy and phenotypic diversity have a strong positive correlation and this relationship is consistent across various spatial configurations. Diversity also increases evolutionary potential, but has a much smaller impact than other components of environmental composition. The most important of these components was the mean number of resources present in locations across the environment, likely owing to the importance of building blocks for the evolution of complex features. These results suggest that a general relationship exists between spatial heterogeneity and diversity, beyond the specific ecosystems and timescales in which it has previously been studied. By examining this relationship in the context of phenotypic evolution, we advance a mechanistic understanding of the resulting dynamics. Moreover, our results suggest that the likelihood of evolving various traits can be impacted by the spatial configuration of patches in which these traits are advantageous. These findings have implications for both evolutionary biology and evolutionary computation, as generating and maintaining diversity is critical to all forms of evolution.},
journal = {bioRxiv},
}
@article {Dufresne2003,
author = {Dufresne, Alexis and Salanoubat, Marcel and Partensky, Fr{\'e}d{\'e}ric and Artiguenave, Fran{\c c}ois and Axmann, Ilka M. and Barbe, Val{\'e}rie and Duprat, Simone and Galperin, Michael Y. and Koonin, Eugene V. and Le Gall, Florence and Makarova, Kira S. and Ostrowski, Martin and Oztas, Sophie and Robert, Catherine and Rogozin, Igor B. and Scanlan, David J. and de Marsac, Nicole Tandeau and Weissenbach, Jean and Wincker, Patrick and Wolf, Yuri I. and Hess, Wolfgang R.},
title = {Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome},
volume = {100},
number = {17},
pages = {10020--10025},
year = {2003},
publisher = {National Academy of Sciences},
journal = {Proceedings of the National Academy of Sciences}
}
@ARTICLE{Durham2013,
author={Durham, W.M. and Climent, E. and Barry, M. and De Lillo, F. and Boffetta, G. and Cencini, M. and Stocker, R.},
title={Turbulence drives microscale patches of motile phytoplankton},
journal={Nature Communications},
year={2013},
volume={4},
art_number={2148},
}
@ARTICLE{Falkowski1998,
author={Falkowski, P.G. and Barber, R.T. and Smetacek, V.},
title={Biogeochemical controls and feedbacks on ocean primary production},
journal={Science},
year={1998},
volume={281},
number={5374},
pages={200-206},
}
@article {Follows2007,
author = {Follows, Michael J. and Dutkiewicz, Stephanie and Grant, Scott and Chisholm, Sallie W.},
title = {Emergent Biogeography of Microbial Communities in a Model Ocean},
volume = {315},
number = {5820},
pages = {1843--1846},
year = {2007},
doi = {10.1126/science.1138544},
publisher = {American Association for the Advancement of Science},
abstract = {A marine ecosystem model seeded with many phytoplankton types, whose physiological traits were randomly assigned from ranges defined by field and laboratory data, generated an emergent community structure and biogeography consistent with observed global phytoplankton distributions. The modeled organisms included types analogous to the marine cyanobacterium Prochlorococcus. Their emergent global distributions and physiological properties simultaneously correspond to observations. This flexible representation of community structure can be used to explore relations between ecosystems, biogeochemical cycles, and climate change.},
issn = {0036-8075},
journal = {Science}
}
@article {Geyrhofer2018,
author = {Geyrhofer, Lukas and Brenner, Naama},
title = {Coexistence and cooperation in structured habitats},
elocation-id = {429605},
year = {2018},
doi = {10.1101/429605},
publisher = {Cold Spring Harbor Laboratory},
journal = {bioRxiv}
}
@ARTICLE{Gonze2018,
author={Gonze, D. and Coyte, K.Z. and Lahti, L. and Faust, K.},
title={Microbial communities as dynamical systems},
journal={Current Opinion in Microbiology},
year={2018},
volume={44},
pages={41-49},
}
@ARTICLE{Goyal2018,
author={Goyal, A. and Maslov, S.},
title={Diversity, Stability, and Reproducibility in Stochastically Assembled Microbial Ecosystems},
journal={Physical Review Letters},
year={2018},
volume={120},
number={15},
art_number={158102},
}
@article{Guirey2010,
title = {Persistence of cluster synchronization under the influence of advection},
author = {Guirey, Emma and Bees, Martin and Martin, Adrian and Srokosz, Meric},
journal = {Phys. Rev. E},
volume = {81},
issue = {5},
pages = {051902},
numpages = {16},
year = {2010},
month = {May},
publisher = {American Physical Society},
}
@ARTICLE{Hallegraeff2010,
author={Hallegraeff, G.M.},
title={Ocean climate change, phytoplankton community responses, and harmful algal blooms: A formidable predictive challenge},
journal={Journal of Phycology},
year={2010},
volume={46},
number={2},
pages={220-235},
}
@ARTICLE{Hardin1960,
author={Hardin, G.},
title={The competitive exclusion principle},
journal={Science},
year={1960},
volume={131},
number={3409},
pages={1292-1297},
}
@Article{Hays2017,
author={Hays, Stephanie G. and Yan, Leo L. W. and Silver, Pamela A. and Ducat, Daniel C.},
title={Synthetic photosynthetic consortia define interactions leading to robustness and photoproduction},
journal={Journal of Biological Engineering},
year={2017},
volume={11},
number={1},
pages={4},
}
@InCollection{Huggett2019,
author = {Huggett, Nick},
title = {Zeno’s Paradoxes},
booktitle = {The Stanford Encyclopedia of Philosophy},
editor = {Edward N. Zalta},
howpublished = {\url{https://plato.stanford.edu/archives/spr2019/entries/paradox-zeno/}},
year = {2019},
edition = {Spring 2019},
publisher = {Metaphysics Research Lab, Stanford University},
}
@article{Hubbell2006,
author = {Hubbell, Stephen P.},
title = {NEUTRAL THEORY AND THE EVOLUTION OF ECOLOGICAL EQUIVALENCE},
journal = {Ecology},
volume = {87},
number = {6},
pages = {1387-1398},
abstract = {Since the publication of the unified neutral theory in 2001, there has been much discussion of the theory, pro and con. The hypothesis of ecological equivalence is the fundamental yet controversial idea behind neutral theory. Assuming trophically similar species are demographically alike (symmetric) on a per capita basis is only an approximation, but it is equivalent to asking: How many of the patterns of ecological communities are the result of species similarities, rather than of species differences? The strategy behind neutral theory is to see how far one can get with the simplification of assuming ecological equivalence before introducing more complexity. In another paper, I review the empirical evidence that led me to hypothesize ecological equivalence among many of the tree species in the species-rich tropical forest on Barro Colorado Island (BCI). In this paper, I develop a simple model for the evolution of ecological equivalence or niche convergence, using as an example evolution of the suite of life history traits characteristic of shade tolerant tropical tree species. Although the model is simple, the conclusions from it seem likely to be robust. I conclude that ecological equivalence for resource use are likely to evolve easily and often, especially in species-rich communities that are dispersal and recruitment limited. In the case of the BCI forest, tree species are strongly dispersal- and recruitment-limited, not only because of restricted seed dispersal, but also because of low recruitment success due to heavy losses of the seedling stages to predators and pathogens and other abiotic stresses such as drought. These factors and the high species richness of the community strongly reduce the potential for competitive exclusion of functionally equivalent or nearly equivalent species.},
year = {2006},
}
@article{Huisman2001,
author = {Huisman, Jef and Johansson, Anna M. and Folmer, Eelke O. and Weissing, Franz J. },
title = {Towards a solution of the plankton paradox: the importance of physiology and life history},
journal = {Ecology Letters},
volume = {4},
number = {5},
pages = {408-411},
abstract = {Phytoplankton communities reveal an astonishing biodiversity, whereas classical competition theory seems to suggest that only a few competing species can survive. Recently we suggested a new solution to this plankton paradox. In theory, at least, competition between multiple species can generate complex dynamics that can support a large number of species. How likely is it then, in reality, that competitive chaos indeed promotes biodiversity? To obtain some insight, we simulated multispecies competition according to five different physiological scenarios. For random species parameters, biodiversity was generally low. Assuming plausible physiological trade-offs, the simulations revealed switches back and forth between equilibrium and nonequilibrium dynamics, and a higher biodiversity. An extremely high biodiversity, with sometimes more than 100 species on three resources, was observed in simulations that assumed a cyclic relation between competitive abilities and resource contents. We conclude that physiological and life-history patterns have a major impact on the likelihood of nonequilibrium dynamics and on the biodiversity of plankton communities.},
year = {2001},
}
@article{Hutchinson1961,
author = {Hutchinson, G. E.},
title = {The Paradox of the Plankton},
journal = {The American Naturalist},
volume = {95},
number = {882},
pages = {137-145},
year = {1961},
}
@article{Hwang2010,
title={Patterns of zooplankton distribution along the marine, estuarine and riverine portions of the Danshuei ecosystem in northern Taiwan},
author={Hwang, Jiang-Shiou and Kumar, Ram and Hsieh, Chih-Wei and Kuo, Albert Y and Souissi, Sami and Hsu, Ming-Hsi and Wu, Jiunn-Tzong and Liu, Wen-Cheng and Wang, Chi-Fang and Chen, Qing-Chao and others},
journal={Zoological Studies},
volume={49},
number={3},
pages={335--352},
year={2010},
}
@article{Irigoien2004,
author={Irigoien, X. and Hulsman, J. and Harris, R.P.},
title={Global biodiversity patterns of marine phytoplankton and zooplankton},
journal={Nature},
year={2004},
volume={429},
number={6994},
pages={863-867},
}
@ARTICLE{Johnson2011,
author={Johnson, K.A. and Goody, R.S.},
title={The original Michaelis constant: Translation of the 1913 Michaelis-Menten Paper},
journal={Biochemistry},
year={2011},
volume={50},
number={39},
pages={8264-8269},
doi={10.1021/bi201284u},
note={cited By 349},
document_type={Article},
source={Scopus},
}
@article{Laan2018,
author = {Andres Laan and Gonzalo G. de Polavieja },
title = {Species diversity rises exponentially with the number of available resources in a multi-trait competition model},
journal = {Proceedings of the Royal Society B: Biological Sciences},
volume = {285},
number = {1885},
pages = {20181273},
year = {2018},
}
@article{Leibold2004,
author = {Leibold, M. A. and Holyoak, M. and Mouquet, N. and Amarasekare, P. and Chase, J. M. and Hoopes, M. F. and Holt, R. D. and Shurin, J. B. and Law, R. and Tilman, D. and Loreau, M. and Gonzalez, A.},
title = {The metacommunity concept: a framework for multi-scale community ecology},
journal = {Ecology Letters},
volume = {7},
number = {7},
pages = {601-613},
keywords = {Mass effects, metacommunity, neutral model, patch dynamics, species sorting},
abstract = {Abstract The metacommunity concept is an important way to think about linkages between different spatial scales in ecology. Here we review current understanding about this concept. We first investigate issues related to its definition as a set of local communities that are linked by dispersal of multiple potentially interacting species. We then identify four paradigms for metacommunities: the patch-dynamic view, the species-sorting view, the mass effects view and the neutral view, that each emphasizes different processes of potential importance in metacommunities. These have somewhat distinct intellectual histories and we discuss elements related to their potential future synthesis. We then use this framework to discuss why the concept is useful in modifying existing ecological thinking and illustrate this with a number of both theoretical and empirical examples. As ecologists strive to understand increasingly complex mechanisms and strive to work across multiple scales of spatio-temporal organization, concepts like the metacommunity can provide important insights that frequently contrast with those that would be obtained with more conventional approaches based on local communities alone.},
year = {2004},
}
@ARTICLE{Lindemann2017,
author={Lindemann, C. and Visser, A. and Mariani, P.},
title={Dynamics of phytoplankton blooms in turbulent vortex cells},
journal={Journal of the Royal Society Interface},
year={2017},
volume={14},
number={136},
}
@ARTICLE{Litchman2008,
author={Litchman, E. and Klausmeier, C.A.},
title={Trait-based community ecology of phytoplankton},
journal={Annual Review of Ecology, Evolution, and Systematics},
year={2008},
volume={39},
pages={615-639},
}
@article {Locey2016,
author = {Locey, Kenneth J. and Lennon, Jay T.},
title = {Scaling laws predict global microbial diversity},
volume = {113},
number = {21},
pages = {5970--5975},
year = {2016},
doi = {10.1073/pnas.1521291113},
publisher = {National Academy of Sciences},
issn = {0027-8424},
journal = {Proceedings of the National Academy of Sciences},
}
@article{Lombard2019,
author={Lombard, Fabien and Boss, Emmanuel and Waite, Anya M. and Vogt, Meike and Uitz, Julia and Stemmann, Lars and Sosik, Heidi M. and Schulz, Jan and Romagnan, Jean-Baptiste and Picheral, Marc and Pearlman, Jay and Ohman, Mark D. and Niehoff, Barbara and Möller, Klas O. and Miloslavich, Patricia and Lara-Lpez, Ana and Kudela, Raphael and Lopes, Rubens M. and Kiko, Rainer and Karp-Boss, Lee and Jaffe, Jules S. and Iversen, Morten H. and Irisson, Jean-Olivier and Fennel, Katja and Hauss, Helena and Guidi, Lionel and Gorsky, Gaby and Giering, Sarah L. C. and Gaube, Peter and Gallager, Scott and Dubelaar, George and Cowen, Robert K. and Carlotti, François and Briseño-Avena, Christian and Berline, Léo and Benoit-Bird, Kelly and Bax, Nicholas and Batten, Sonia and Ayata, Sakina Dorothée and Artigas, Luis Felipe and Appeltans, Ward},
title={Globally Consistent Quantitative Observations of Planktonic Ecosystems},
journal={Frontiers in Marine Science},
volume={6},
pages={196},
year={2019},
}
@article{Loreau2003,
author = {Loreau, Michel and Mouquet, Nicolas and Holt, Robert D.},
title = {Meta-ecosystems: a theoretical framework for a spatial ecosystem ecology},
journal = {Ecology Letters},
volume = {6},
number = {8},
pages = {673-679},
keywords = {Dispersal, diversity, ecosystem, landscape, metacommunity, model, productivity, source–sink dynamics, spatial processes},
abstract = {Abstract This contribution proposes the meta-ecosystem concept as a natural extension of the metapopulation and metacommunity concepts. A meta-ecosystem is defined as a set of ecosystems connected by spatial flows of energy, materials and organisms across ecosystem boundaries. This concept provides a powerful theoretical tool to understand the emergent properties that arise from spatial coupling of local ecosystems, such as global source–sink constraints, diversity–productivity patterns, stabilization of ecosystem processes and indirect interactions at landscape or regional scales. The meta-ecosystem perspective thereby has the potential to integrate the perspectives of community and landscape ecology, to provide novel fundamental insights into the dynamics and functioning of ecosystems from local to global scales, and to increase our ability to predict the consequences of land-use changes on biodiversity and the provision of ecosystem services to human societies.},
year = {2003},
}
@ARTICLE{Lowery2019,
author={Lowery, N.V. and Ursell, T.},
title={Structured environments fundamentally alter dynamics and stability of ecological communities},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2019},
volume={116},
number={2},
pages={379-388},
}
@ARTICLE{Macias2010,
author={Macías, D. and Somavilla, R. and González-Gordillo, J.I. and Echevarría, F.},
title={Physical control of zooplankton distribution at the Strait of Gibraltar during an episode of internal wave generation},
journal={Marine Ecology Progress Series},
year={2010},
volume={408},
pages={79-85},
}
@ARTICLE{Malviya2016,
author={Malviya, S. and Scalco, E. and Audic, S. and Vincent, F. and Veluchamy, A. and Poulain, J. and Wincker, P. and Iudicone, D. and De Vargas, C. and Bittner, L. and Zingone, A. and Bowler, C.},
title={Insights into global diatom distribution and diversity in the world's ocean},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2016},
volume={113},
number={11},
pages={E1516-E1525},
}
@article{Margalef1994,
author = {Margalef, R},
year = {1994},
month = {01},
pages = {87-101},
title = {Through the looking glass: How marine phytoplankton appears through the microscope when graded by size and taxonomically sorted},
volume = {58},
journal = {Scientia Marina}
}
@ARTICLE{Marsland2019,
author={Marsland, R., III and Cui, W. and Goldford, J. and Sanchez, A. and Korolev, K. and Mehta, P.},
title={Available energy fluxes drive a transition in the diversity, stability, and functional structure of microbial communities},
journal={PLoS Computational Biology},
year={2019},
volume={15},
number={2},
}
@article{Mora2011,
author = {Mora, Camilo AND Tittensor, Derek P. AND Adl, Sina AND Simpson, Alastair G. B. AND Worm, Boris},
journal = {PLOS Biology},
publisher = {Public Library of Science},
title = {How Many Species Are There on Earth and in the Ocean?},
year = {2011},
month = {08},
volume = {9},
pages = {1-8},
number = {8},
}
@ARTICLE{Niehaus2019,
author={Niehaus, L. and Boland, I. and Liu, M. and Chen, K. and Fu, D. and Henckel, C. and Chaung, K. and Miranda, S.E. and Dyckman, S. and Crum, M. and Dedrick, S. and Shou, W. and Momeni, B.},
title={Microbial coexistence through chemical-mediated interactions},
journal={Nature Communications},
year={2019},
volume={10},
number={1},
art_number={2052},
}
@article{Ohman2019,
author = {Ohman, Mark D. and Davis, Russ E. and Sherman, Jeffrey T. and Grindley, Kyle R. and Whitmore, Benjamin M. and Nickels, Catherine F. and Ellen, Jeffrey S.},
title = {Zooglider: An autonomous vehicle for optical and acoustic sensing of zooplankton},
journal = {Limnology and Oceanography: Methods},
volume = {17},
number = {1},
pages = {69-86},
doi = {10.1002/lom3.10301},
abstract = {Abstract We present the design and preliminary results from ocean deployments of Zooglider, a new autonomous zooplankton-sensing glider. Zooglider is a modified Spray glider that includes a low-power camera (Zoocam) with telecentric lens and a custom dual frequency Zonar (200 and 1000 kHz). The Zoocam quantifies zooplankton and marine snow as they flow through a defined volume inside a sampling tunnel. Images are acquired on average every 5 cm from a maximum operating depth of ~ 400 m to the sea surface. Biofouling is mitigated using a dual approach: an ultraviolet light-emitting diode and a mechanical wiper. The Zonar permits differentiation of large and small acoustic backscatterers in larger volumes than can be sampled optically. Other sensors include a pumped conductivity, temperature, and depth unit and chlorophyll a fluorometer. Zooglider enables fully autonomous in situ measurements of mesozooplankton distributions, together with the three-dimensional orientation of organisms and marine snow in relation to other biotic and physical properties of the ocean water column. It is well suited to resolve thin layers and microscale ocean patchiness. Battery capacity supports 50 d of operations. Zooglider includes two-way communications via Iridium, permitting near-real–time transmission of data from each dive profile, as well as interactive instrument control from remote locations for adaptive sampling.},
year = {2019},
}
@ARTICLE{Pesant2015,
author={Pesant, S. and Not, F. and Picheral, M. and Kandels-Lewis, S. and Le Bescot, N. and Gorsky, G. and Iudicone, D. and Karsenti, E. and Speich, S. and Trouble, R. and Dimier, C. and Searson, S.},
title={Open science resources for the discovery and analysis of Tara Oceans data},
journal={Scientific Data},
year={2015},
volume={2},
art_number={150023},
}
@article{Perez2007,
title = "Phytoplankton absorption spectra along the water column in deep North Patagonian Andean lakes (Argentina)",
journal = "Limnologica",
volume = "37",
number = "1",
pages = "3 - 16",
year = "2007",
note = "Limnology of Temperate South America",
issn = "0075-9511",
author = "Gonzalo Pérez and Claudia Queimaliños and Esteban Balseiro and Beatriz Modenutti",
}
@ARTICLE{Posfai2017,
author={Posfai, A. and Taillefumier, T. and Wingreen, N.S.},
title={Metabolic Trade-Offs Promote Diversity in a Model Ecosystem},
journal={Physical Review Letters},
year={2017},
volume={118},
number={2},
}
@article{Prairie2012,
author = {Prairie, Jennifer C. and Sutherland, Kelly R. and Nickols, Kerry J. and Kaltenberg, Amanda M.},
title = {Biophysical interactions in the plankton: A cross-scale review},
journal = {Limnology and Oceanography: Fluids and Environments},
volume = {2},
number = {1},
pages = {121-145},
keywords = {plankton distributions, plankton dynamics, turbulence, biomixing, interdisciplinary, coupling, technology},
abstract = {Lay Abstract The study of plankton ecology is by nature interdisciplinary, since both biology and physics interact to shape plankton distributions and population dynamics. Small-scale turbulence affects how plankton feed and grow, while kilometer-scale physical features can form large plankton patches. Recent findings also show that, in addition to physics affecting plankton dynamics, plankton may be able to influence ocean physics as well. For example, the fluid motion produced from a copepod swimming in pursuit of food can be detected by nearby organisms and might make it more vulnerable to its predators. The field of plankton biophysics has advanced rapidly in the past several decades, due in large part to technological advances that allow the study of interactions between fluid flow and plankton in their natural environment at smaller scales than ever before. Biophysical interactions in plankton ecology can occur at a wide range of scales: from the scale of individual organisms up to the scale of the world's oceans. In this article, we review biophysical interactions in plankton across a wide range of scales, focusing on the most recent research in the field. In addition, we discuss how these processes are linked with other processes that occur at smaller and larger scales. For example, plankton patches formed by ocean eddies can provide feeding grounds for other organisms, thus influencing global ocean food web dynamics. Finally, we discuss some of the potential effects of long-term changes in global climate, which will affect ocean temperature and stratification, thus regulating the occurrence and intensity of biophysical plankton processes.},
year = {2012},
}
@article{Preston1962,
author = {Preston, F. W.},
title = {The Canonical Distribution of Commonness and Rarity: Part I},
journal = {Ecology},
volume = {43},
number = {2},
pages = {185-215},
year = {1962},
}
@Book{Quine1976,
author = { Quine, W. V. },
title = { The ways of paradox, and other essays},
edition = { Rev. and enl. ed. },
isbn = { 0674948351 0674948378 },
publisher = { Harvard University Press Cambridge, Mass },
pages = { x, 335 p. ; },
year = { 1976 },
type = { Book },
}
@ARTICLE{Reichenbach2006,
author={Reichenbach, T. and Mobilia, M. and Frey, E.},
title={Coexistence versus extinction in the stochastic cyclic Lotka-Volterra model},
journal={Physical Review E - Statistical, Nonlinear, and Soft Matter Physics},
year={2006},
volume={74},
number={5},
}
@book{Reynolds2006,
place={Cambridge},
series={Ecology, Biodiversity and Conservation},
title={The Ecology of Phytoplankton},
DOI={10.1017/CBO9780511542145},
publisher={Cambridge University Press},
author={Reynolds, C. S.},
year={2006},
collection={Ecology, Biodiversity and Conservation}}
@article {Ring1981,
author={The Ring Group},
title = {Gulf Stream Cold-Core Rings: Their Physics, Chemistry, and Biology},
volume = {212},
number = {4499},
pages = {1091--1100},
year = {1981},
publisher = {American Association for the Advancement of Science},
issn = {0036-8075},
journal = {Science}
}
@ARTICLE{Roeselers2007,
author={Roeselers, G. and Norris, T.B. and Castenholz, R.W. and Rysgaard, S. and Glud, R.N. and Kühl, M. and Muyzer, G.},
title={Diversity of phototrophic bacteria in microbial mats from Arctic hot springs (Greenland)},
journal={Environmental Microbiology},
year={2007},
volume={9},
number={1},
pages={26-38},
}
@Article{Rescigno1965,
author={Rescigno, Aldo and Richardson, Irvin W.},
title={On the competitive exclusion principle},
journal={The bulletin of mathematical biophysics},
year={1965},
month={Jan},
volume={27},
number={1},
pages={85-89},
}
@book{Sainsbury1995,
place={Cambridge},
edition={2},
title={Paradoxes},
publisher={Cambridge University Press},
author={Sainsbury, R. M.},
year={1995},
}
@Article{Scheffer2003,
author="Scheffer, Marten
and Rinaldi, Sergio
and Huisman, Jef
and Weissing, Franz J.",
title="Why plankton communities have no equilibrium: solutions to the paradox",
journal="Hydrobiologia",
year="2003",
month="Jan",
day="01",
volume="491",
number="1",
pages="9--18",
}
@ARTICLE{Sengupta2017,
author={Sengupta, A. and Carrara, F. and Stocker, R.},
title={Phytoplankton can actively diversify their migration strategy in response to turbulent cues},
journal={Nature},
year={2017},
volume={543},
number={7646},
pages={555-558},
}
@article {Shoval2012,
author = {Shoval, O. and Sheftel, H. and Shinar, G. and Hart, Y. and Ramote, O. and Mayo, A. and Dekel, E. and Kavanagh, K. and Alon, U.},
title = {Evolutionary Trade-Offs, Pareto Optimality, and the Geometry of Phenotype Space},
volume = {336},
number = {6085},
pages = {1157--1160},
year = {2012},
publisher = {American Association for the Advancement of Science},
abstract = {Biological systems that perform multiple tasks face a fundamental trade-off: A given phenotype cannot be optimal at all tasks. Here we ask how trade-offs affect the range of phenotypes found in nature. Using the Pareto front concept from economics and engineering, we find that best{\textendash}trade-off phenotypes are weighted averages of archetypes{\textemdash}phenotypes specialized for single tasks. For two tasks, phenotypes fall on the line connecting the two archetypes, which could explain linear trait correlations, allometric relationships, as well as bacterial gene-expression patterns. For three tasks, phenotypes fall within a triangle in phenotype space, whose vertices are the archetypes, as evident in morphological studies, including on Darwin{\textquoteright}s finches. Tasks can be inferred from measured phenotypes based on the behavior of organisms nearest the archetypes.},
issn = {0036-8075},
journal = {Science},
}
@ARTICLE{Smith2005,
author={Smith, V.H. and Foster, B.L. and Grover, J.P. and Holt, R.D. and Leibold, M.A. and DeNoyelles Jr., F.},
title={Phytoplankton species richness scales consistently from laboratory microcosms to the world's oceans},
journal={Proceedings of the National Academy of Sciences of the United States of America},
year={2005},
volume={102},
number={12},
pages={4393-4396},
}
@ARTICLE{Smith2016,
author={Smith, S.L. and Pahlow, M. and Merico, A. and Acevedo-Trejos, E. and Sasai, Y. and Yoshikawa, C. and Sasaoka, K. and Fujiki, T. and Matsumoto, K. and Honda, M.C.},
title={Flexible phytoplankton functional type (FlexPFT) model: Size-scaling of traits and optimal growth},
journal={Journal of Plankton Research},
year={2016},
volume={38},
number={4},
pages={977-992},
}
@article {Smriga2016,
author = {Smriga, Steven and Fernandez, Vicente I. and Mitchell, James G. and Stocker, Roman},
title = {Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria},
volume = {113},
number = {6},
pages = {1576--1581},
year = {2016},
publisher = {National Academy of Sciences},
abstract = {Microscale interactions between bacteria and phytoplankton underpin ocean biogeochemistry and frequently involve bacterial chemotaxis to phytoplankton dissolved organic matter (DOM). Yet, it remains unclear how the effects of this interaction propagate to ecosystem scales. We address this gap through a hybrid approach where high-resolution observations of chemotaxis toward a diatom are directly used in a resource utilization model. We find that chemotactic bacteria consume most diatom DOM under resource-rich or bacteria-rich conditions, that DOM is partitioned among distinct populations based on diffusivity, and that consumption is skewed toward very few cells. Nonmotile oligotrophic bacteria dominate when productivity is low. Motile copiotrophs dominate during blooms. Ocean chemotaxis thus partitions resources spatially, by molecular size, and temporally through seasonal and episodic blooms.The microenvironment surrounding individual phytoplankton cells is often rich in dissolved organic matter (DOM), which can attract bacteria by chemotaxis. These {\textquotedblleft}phycospheres{\textquotedblright} may be prominent sources of resource heterogeneity in the ocean, affecting the growth of bacterial populations and the fate of DOM. However, these effects remain poorly quantified due to a lack of quantitative ecological frameworks. Here, we used video microscopy to dissect with unprecedented resolution the chemotactic accumulation of marine bacteria around individual Chaetoceros affinis diatoms undergoing lysis. The observed spatiotemporal distribution of bacteria was used in a resource utilization model to map the conditions under which competition between different bacterial groups favors chemotaxis. The model predicts that chemotactic, copiotrophic populations outcompete nonmotile, oligotrophic populations during diatom blooms and bloom collapse conditions, resulting in an increase in the ratio of motile to nonmotile cells and in the succession of populations. Partitioning of DOM between the two populations is strongly dependent on the overall concentration of bacteria and the diffusivity of different DOM substances, and within each population, the growth benefit from phycospheres is experienced by only a small fraction of cells. By informing a DOM utilization model with highly resolved behavioral data, the hybrid approach used here represents a new path toward the elusive goal of predicting the consequences of microscale interactions in the ocean.},
issn = {0027-8424},
journal = {Proceedings of the National Academy of Sciences},
}
@article{Stein2014,
author = {Stein, Anke and Gerstner, Katharina and Kreft, Holger},
title = {Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales},
journal = {Ecology Letters},
volume = {17},
number = {7},
pages = {866-880},
keywords = {Habitat diversity, habitat structure, meta-analysis, meta-regression, robust variance estimation, spatial scale, species diversity, topographical heterogeneity, vegetation structure},
abstract = {Abstract Environmental heterogeneity is regarded as one of the most important factors governing species richness gradients. An increase in available niche space, provision of refuges and opportunities for isolation and divergent adaptation are thought to enhance species coexistence, persistence and diversification. However, the extent and generality of positive heterogeneity–richness relationships are still debated. Apart from widespread evidence supporting positive relationships, negative and hump-shaped relationships have also been reported. In a meta-analysis of 1148 data points from 192 studies worldwide, we examine the strength and direction of the relationship between spatial environmental heterogeneity and species richness of terrestrial plants and animals. We find that separate effects of heterogeneity in land cover, vegetation, climate, soil and topography are significantly positive, with vegetation and topographic heterogeneity showing particularly strong associations with species richness. The use of equal-area study units, spatial grain and spatial extent emerge as key factors influencing the strength of heterogeneity–richness relationships, highlighting the pervasive influence of spatial scale in heterogeneity–richness studies. We provide the first quantitative support for the generality of positive heterogeneity–richness relationships across heterogeneity components, habitat types, taxa and spatial scales from landscape to global extents, and identify specific needs for future comparative heterogeneity–richness research.},
year = {2014},
}
@article{Taylor1990,
author = {R. Taylor, L and T. A. Picket, S and White, Peter},
year = {1990},
month = {10},
pages = {1199},
title = {The Ecology of Natural Disturbance as Patch Dynamics},
volume = {59},
journal = {The Journal of Animal Ecology},
}
@article{Tian2013,
title = {Delay-driven irregular spatiotemporal patterns in a plankton system},
author = {Tian, Canrong and Zhang, Lai},
journal = {Phys. Rev. E},
volume = {88},
issue = {1},
pages = {012713},
numpages = {6},
year = {2013},
month = {Jul},
publisher = {American Physical Society},
}
@ARTICLE{Turnbaugh2006,
author={Turnbaugh, P.J. and Ley, R.E. and Mahowald, M.A. and Magrini, V. and Mardis, E.R. and Gordon, J.I.},
title={An obesity-associated gut microbiome with increased capacity for energy harvest},
journal={Nature},
year={2006},
volume={444},
number={7122},
pages={1027-1031},
}
@ARTICLE{Venkata2016,
author={Venkata Mohan, S. and Modestra, J.A. and Amulya, K. and Butti, S.K. and Velvizhi, G.},
title={A Circular Bioeconomy with Biobased Products from CO 2 Sequestration},
journal={Trends in Biotechnology},
year={2016},
volume={34},
number={6},
pages={506-519},
}
@article{Volterra1928,
author = {Volterra, Vito},
title = "{Variations and Fluctuations of the Number of Individuals in Animal Species Living Together}",
journal = {ICES Journal of Marine Science},
volume = {3},
number = {1},
pages = {3-51},
year = {1928},
month = {04},
issn = {1054-3139},
}
@article{Wilson1992,
author = {Wilson, David Sloan},
title = {Complex Interactions in Metacommunities, with Implications for Biodiversity and Higher Levels of Selection},
journal = {Ecology},
volume = {73},
number = {6},
pages = {1984-2000},
doi = {10.2307/1941449},
abstract = {Two common features of biological communities are (a) complex interactions among species, which make community dynamics sensitive to initial conditions, and (b) spatial heterogeneity, which fragments large—scale ecological systems into a mosaic of patches, hereafter termed a \say{metacommunity}. This computer simulation study examines the effect of complex interaction on the global and local dynamics of metacommunities. Patches are physically identical and differ only in the initial proportion of species that colonize the patches. The random variation is then magnified by deterministic interactions that cause patches to follow different trajectories based on initial conditions. After a period of interaction, individuals from all patches join in global pool of dispersers that colonize a new 'generation' of patches. Complex interactions can have at least two important effects on metacommunity dynamics. First the number of species coexisting in the metacommunity can greatly exceed the number of species coexisting in any single patch, despite the fact that the patches are physically identical, the species do not differ in colonization ability, and stochastic effects are absent after the colonization stage. Second, complex interactions provide a new source of variation upon which natural selection can operate at the patch level, providing a mechanism for the evolution of functionally organized communities.},
year = {1992},
}
@Book{Whittaker1965,
author = {Whittaker, Robert H. },
title = { Communities and Ecosystems},
publisher = { Macmillan },
pages = { xi, 162 p. },
year = { 1970 },
type = { Book },
}
@article {Woese2004,
author = {Woese, Carl R.},
title = {A New Biology for a New Century},
volume = {68},
number = {2},
pages = {173--186},
year = {2004},
publisher = {American Society for Microbiology Journals},
issn = {1092-2172},
journal = {Microbiology and Molecular Biology Reviews},
}
@ARTICLE{Zhang2019,
author={Zhang, W. and Ding, W. and Li, Y.-X. and Tam, C. and Bougouffa, S. and Wang, R. and Pei, B. and Chiang, H. and Leung, P. and Lu, Y. and Sun, J. and Fu, H. and Bajic, V.B. and Liu, H. and Webster, N.S. and Qian, P.-Y.},
title={Marine biofilms constitute a bank of hidden microbial diversity and functional potential},
journal={Nature Communications},
year={2019},
volume={10},
number={1},
art_number={517},
}