This is an archive of essays, protocols, scripts, lecture notes, practicals and other resources
Calculators
Lab protocols and calculators
-
GoldenGate Calculator Online calculator for DNA Assembly.
Code
MATLAB scripts
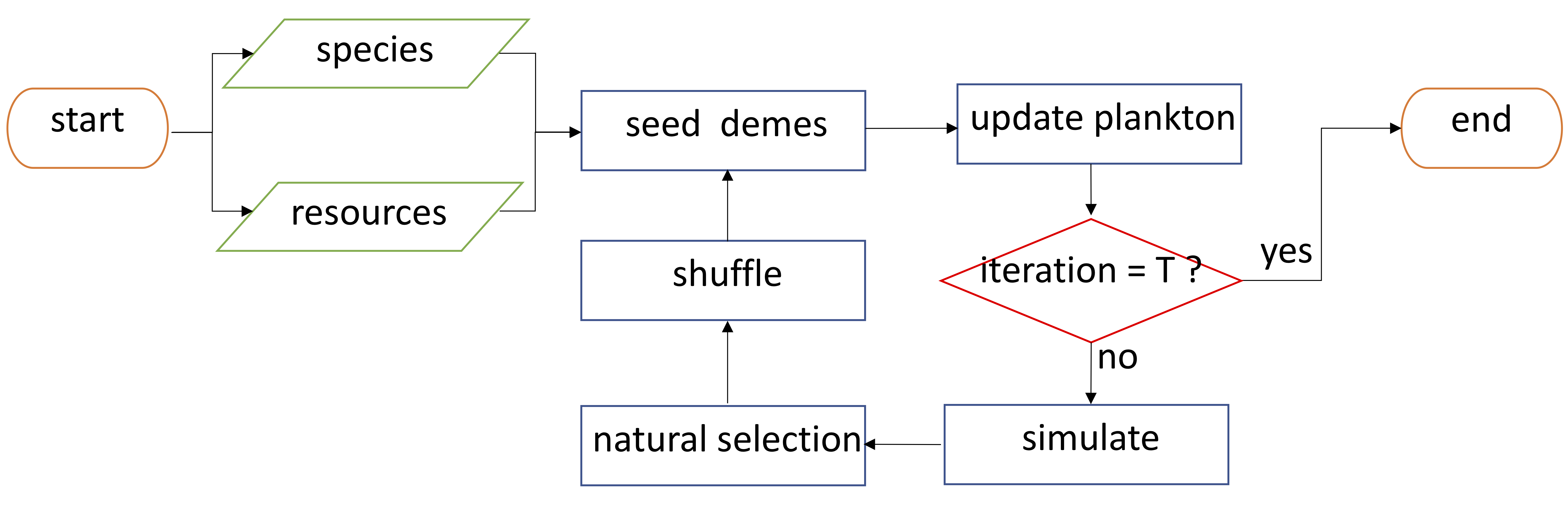
- Plankton Simulator The plankton-simulator is a collection of matlab scripts simulating the dynamics of structured microbial communities. The simulator is based on a multiscale biophysical model that incorporates species traits, consumer-resource dynamics and stochastic ecosystem assembly. The model hierarchically simulates features of ecosystem at different scales by defining three classes in the ecosystem using object-oriented programming in MATLAB: Species, Demes (spatially-isolated subcompartments) and Plankton (collection of Demes).
- Biofilmer
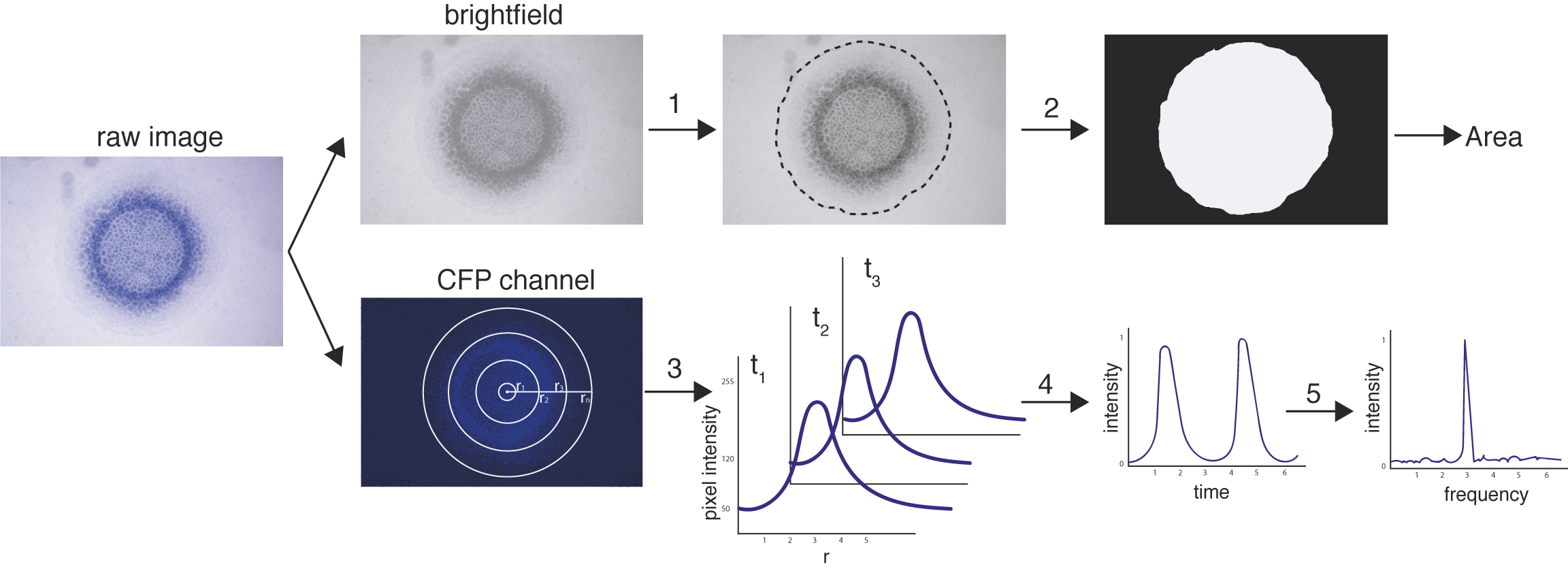
The biofilmer is a repository of MATLAB scripts to quantify properties of fluorescence time-lapse microscopy biofilm images.
Python scripts
-
Plot circular genomes with pyCirclize A repo to store scripts to analyse and visualise results from WGS and Sanger sequencing experiments The scripts are based on python and can be executed via the terminal after having cloned the repo. Many of the circular plot diagrams are made using the package pyCirclize.
Plot Whole Genome Sequencing Results against a Reference Genome
This Python script allows to generate a circular plot of a genomic structure. It draws a diagram of a genome along with various genomic annotations like Forward CDS, Reverse CDS, rRNA, tRNA, and also includes mutation and coverage data for a particular DNA sequencing strain.
Instructions
Prerequisites
Before you run this script, you need to have the following Python packages installed:
- pycirclize
- pandas
- numpy
- matplotlib
- argparse
You can install these using pip:
pip install pycirclize pandas numpy matplotlib argparse
Data Requirements
Your data files should be located in the following paths:
- Coverage data and Identity data:
"Data/reads_alignments/{Strain}_{Reference}.csv" - Mutation data:
"Data/variant_analysis/{Reference}_allmutations.csv" - Genbank file for genome annotations:
"Data/genome_annotations/{Reference}.gff"
Where:
{Strain}is the strain from which the DNA sequencing reads are obtained.{Reference}is the accession number of the reference genome sequencing the reads have been aligned to.
The generated figure will be saved to "Figures/WGS/{Strain}_vs_{Reference}_mutations.png".
Execution
You can run the script using Python3. From the terminal, navigate to the directory containing the script, and run the following command:
python3 GenSeqRefViewer.py -strain= "wt_Nixon" -ref= "NC000911"
Replace <strain_name> with the strain name (as labelled in the csv files) and <reference_name> with the reference genome NCBI accession number (e.g., ‘NC000911’).
Outputs
The script generates a circular diagram of a genome with the given strain and reference genome, highlighting areas of low coverage and unconserved genomic regions. The final diagram is saved as a PNG image with a high resolution (DPI=900). The filename will be of the form "{Strain}_vs_{Reference}_mutations.png" and it will be saved in the "Figures/WGS/" directory.

Text
Old texts and manuscripts
-
Aristotle, Historia Animalium 9.36.20, translated by D’Arcy Wentworth Thompson, 1910
-
ISAAC NEWTON’S GENERAL SCHOLIUM TO THE PRINCIPIA (1713, 1726)
Essays and lecture notes
Genetics, molecular, synthetic biology and biotechnology
- Molecular Biology
- GMO Papayas
- Plant Biotechnology
- Bacterial Genetics
- Genes and Genomes
- Genomic Manipulation
- Molecular Biology of The Chloroplast, Peter Nixon
- Systems Biology
- Biophysics, Robert Endres
- Biophysics, Robert Endres
- Dynamics in Gene Regulation, Robert Endres
- Mathematical Modelling in Biology
- Quantitative imaging of cell topology
- Synthetic Biology
- Synthetic Genomics, Tom Ellis
- Bacterial Logic Gates, Tom Ellis
- Part Libraries in SynBio, Tom Ellis
- Optogenetics, Mark Isalan
- Advanced Synthetic Biology Applications: Pattern Formation, Mark Isalan
- Synthetic Biology IC Exam 15-16
Biochemistry
- Structure of the Nicotine Receptor
- Neurobiology
- Proteins and Enzymes
- Enzymology 1
- Enzymology 2
- DNA Repair
- Bacterial Genetics
- Tryptophan Operon in B.subtilis
- Bioinformatics
- Proteomics
- Protein Folding
- Biological NMR Spectroscopy
- Cryo-EM
Albi’s essays
- Prospects for the Development of Cyanobacterial Biofilm Bioreactors
- Human Practices in Synthetic Biology
- Economics Thought in Italy Before WWII
- Ricordi d’Africa: La Seconda Battaglia di El Alamein